Measuring the Power of Product Features to Generate Increased DemandProduct management requires more from feature prioritization than a rank ordering. It is simply not enough to know the "best" feature if that best does not generate increased demand. We are not searching for the optimal features in order to design a product that no one will buy. We want to create purchase interest; therefore, that is what we ought to measure.
A grounded theory of measurement mimics the purchase process by asking consumers to imagine themselves buying a product. When we wish to assess the impact of adding features, we think first of a choice model where a complete configuration of product features are systematically manipulated according to an experimental design. However, given that we are likely to be testing a considerable number of separate features rather than configuring a product, our task becomes more of a screening process than a product design. Choice modeling requires more than we want to provide, but we would still like customer input concerning the value of each of many individual features.
We would like to know the likely impact that each feature would have if it were added to the product one at a time. Is the feature so valuable that customers will be willing to pay more? Perhaps this is not the case, but could the additional feature serve as a tie-breaker? That is, given a choice between one offer with the feature and another offer without the feature, would the feature determine the choice? If not, does the feature generate any interest or additional appeal? Or, is the feature simply ignored so that the customer reports "not interested" in this feature?
What I am suggesting is an ordinal, behaviorally-anchored scale with four levels: 1=not interested, 2=nice to have, 3=tie-breaker and 4=pay more for. However, I have no commitment to these or any other specific anchors. They are simply one possibility that seems to capture important milestones in the purchase process. What is important is that we ask about features within a realistic purchase context so that the respondent can easily imagine the likely impact of adding each feature to an existing product.
Data collection, then, is simply asking consumers who are likely to purchase the product to evaluate each feature separately and place it into one of the ordered behavioral categories. Feature prioritization is achieved by comparing the impact of the features. For example, if management wants to raise its prices, then only those features falling into the "pay more for" category would be considered a positive response. But we can do better than this piecemeal category-specific comparison by studying the pattern of impact across all the features using item response theory (IRT). In addition, it is important to note that in contrast with importance ratings, a behaviorally anchored scale is directly interpretable. Unlike ratings of "somewhat important" or a four on a five-point scale, the product manager has a clear idea of the likely impact of a "tie-breaker" on purchase interest.
It should be noted that the respondent is never asked to rank order the features or select the best and worst from a set of a four or five features. Feature prioritization is the job of the product manager and not the consumer. Although there are times when incompatible features require the consumer to make tradeoffs in the marketplace (e.g., price vs. quality), this does not occur for most features and is a task with which consumers have little familiarity. Those of us who like sweet-and-sour food have trouble deciding whether we value sweet more than sour. If the feature is concrete, the consumer can react to the feature concept and report its likely impact on their behavior (as long as we are very careful not to infer too much form self-reports). One could argue that such a task is grounded in consumer experience. On the other hand, what marketplace experience mimics the task of selecting the best or the worst among small groupings of different features? Let the consumer react, and let product management do the feature prioritization.
What is Feature Prioritization?
The product manager needs to make a design decision. With limited resources, what is the first feature that should be added? So why not ask for a complete ranking of all the features? For a single person, this question can be answered with a simple ordering of the features. That is, if my goal were to encourage this one person to become more interested in my product, a rank ordering of the features would provide an answer. Why not repeat the same ranking process for n respondents and calculate some type of aggregate rank?
Unfortunately, rank is not purchase likelihood. Consider the following three respondents and their purchase likelihoods for three versions of the same product differing only in what additional feature is included.
| Purchase Likelihood | Rank Ordering |
| f1 | f2 | f3 | f1 | f2 | f3 |
r1 | 0.10 | 0.20 | 0.30 | 3 | 2 | 1 |
r2 | 0.10 | 0.20 | 0.30 | 3 | 2 | 1 |
r3 | 0.90 | 0.50 | 0.10 | 1 | 2 | 3 |
| 0.37 | 0.30 | 0.23 | 2.33 | 2.00 | 1.67 |
What if we had only the rankings and did not know the corresponding purchase interest? Feature 3 is ranked first most often and has the highest average ranking. Yet, unknown to us because we did not collect the data, the highest average purchase likelihood belongs to Feature 1. It appears that we may have ranked prematurely and allowed our desire for feature differentiation to blind us to the unintended consequences.
We, on the other hand, have not taken the bait and have tested the behaviorally-anchored impact of each feature. Consequently, we have a score from 1 to 4 for every respondent on every feature. If there were 200 consumers in the study and 9 features, then we would have a 200 x 9 data matrix filled with 1's, 2's, 3's, and 4's. And now what shall we do?
Analysis of the Behaviorally-Anchored Categories of Feature ImpactFirst, we recognize that our scale values are ordinal. Our consumers read each feature description and ask themselves which category best describes the feature's impact. The behavioral categories are selected to represent milestones in the purchase process. We encourage consumers to think about the feature in the context of marketplace decisions. Our interest is not the value of a feature in some Platonic ideal world, but the feature's actual impact on real-world behavior that will affect the bottom line. When I ask for an importance rating without specifying a context, respondents are on their own with little guidance or constraint. Behaviorally-anchored response categories constrain the respondent to access only the information that is relevant to the task of deciding which of the scale values most accurately represents their intention.
To help understand this point, one can think of the difference between measuring age in equal yearly increments and measuring age in terms of transitional points in the United States such as 18, 21, and 65. In our case we want a series of categories that both span the range of possible feature impacts and have high imagery so that the measurement is grounded in a realistic context. Moreover, we want to be certain to measure the higher ends of feature impact because we are likely to be asking consumers about features that we believe they want (e.g., "pay more for" or "must have" or "first thing I would look for"). Such breakpoints are at best ordinal and require something like the graded response model from item response theory (an overview is available at this
prior post).
Feature prioritization seeks a sequential ordering of the features along a single dimension representing its likely impact on consumers. The consumer acts as a self-informant and reports how they would behave if the feature were added to the product. It is important to note that both the features and consumers vary along the same continuum. Features have greater or less impact, and consumers report varying levels of feature impact. That is, consumer have different levels of involvement with the product so that their wants and needs have different intensity. Greater product involvement leads to higher impact ratings across all the features. For example, cell phone users vary in the intensity with which they use their phone as a camera. As a result, when asked about the likely impact of additional cell phone features associated with the picture quality or camera ease of use, the more intense camera users are likely to give uniformly higher ratings to all the features. The features still receive different scores with the most desirable features getting the highest scores. Consequently, we might expect to observe the same pattern of high and low impact ratings for all the users with elevation of the curve dependent on feature usage intensity.
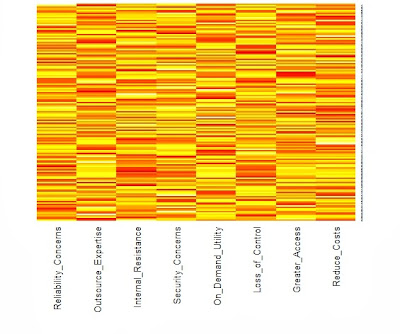
We need an example to illustrate these points. The R package psych contains a function for simulating graded response data. As shown in the appendix, we have random generated 200 respondents who rated 9 features on the 4-point anchored scale previous defined. The heatmap below reveals the underlying structure.
![]()
The categories are represented by color with dark blue=4, light blue=3, light red=2 and dark red=1. The respondents have been sorted by their total scores so that the consistently lowest ratings are indicated by rows of dark red toward the top of the heatmap. As one moves down the heatmap, the rows change gradually from predominantly red to blue. In addition, the columns have been sorted from the features with the lowest average rating to those features with the highest rating. As a result, the color changes in the rows follow a pattern with the most wanted features turning colors first as we proceed down the heatmap. V9 represents a feature that most want, but V1 is desired by only the most intense user. Put another way, our user in the bottom row is a "cell phone picture taking machine" who has to have every feature, even those features with which most others have little interest. However, the users near the top of our heatmap are not interested in any of the additional features.
Multiple Correspondence Analysis (MCA)
In the
prior post that I have already referenced, the reader can find a worked example of how to code and interpret a graded response model using the R package ltm for latent trait modeling. Instead of repeating that analysis with a new data set, I wanted to supplement the graphic displays from heatmaps with the graphic displays from MCA.
Warrens and Heiser have written an overview of this approach, but let me emphasize a couple of points from that article. MCA is a dual scaling technique. Dual scaling refers to the placement of rows and column on the same space. Similar rows are placed near each other and away from dissimilar rows. The same is true for columns. The relationship between rows and columns, however, is not directly plotted on the map, although in general one will find respondents located in the same region as the columns they tended to select. Even if you have had no prior experience with MCA, you should be able to follow the discussion below as if it were nothing more than a description of some scatterplots.
As mentioned before, the R code for all the analysis can be found in the appendix at the end of this post. Unlike a graded response model, MCA treats all category levels as nominal or factors in R. Thus, the 200 x 9 data matrix must be expanded to repeat the four category levels for each of the nine features. That is, a "3" (indicating that the feature would break a tie between two otherwise equivalent products) is represented in MCA by four category levels taking only zero and one values (in this case 0, 0, 1, 0). This means that the 200 x 9 data matrix becomes a 200 x 36 indicator matrix with four columns for each feature.
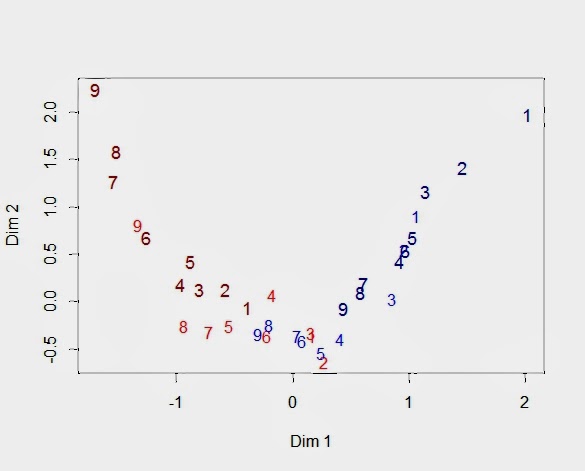
Looking at our heatmap above, the bottom rows with many 3's and 4's (blue) across all the features will be positioned near each other because their response patterns are similar. We would expect the same for the top rows, although in this case it is because the respondents share many 1's and 2's (dark red). We can ask the same location question about the 36 columns, but now the heatmap shows only the relationships among the 9 features. Obviously, adjacent columns are more similar, but we will need to examine the MCA map to learn about the locations of the category levels for each feature. I have presented such a map below displaying the positions of the 36 columns on the first two latent dimensions.
![]()
The numbers refer to the nine features (V1 to V9). There are four 9's because there are four category levels for V9. I used the same color scheme as the heatmap, so that the 1's are dark red, and I made them slightly larger in order differentiate them from the 2's that are a lighter shade of red. Similarly, the light blue numbers refer to category level 3 for each feature, and the slightly larger dark blue numbers are the feature's fourth category (pay more for).
You can think of this "arc" as a path along which the rows of the heatmap fall. As you move along this path from the upper left to the upper right, you will trace out the rows of the heatmap. That is, the dark red "9" indicates the lowest score for Feature 9. It is followed by the lowest score for Feature 8 and Feature 7. What comes next? The light red for Feature 9 indicates the second lowest score for Feature 9. Then we see the lowest scores for the remaining features. This is the top of our heatmap. When we get to the end of our path, we see a similar pattern in reverse with respondents at the bottom of the heatmap.
Sometimes it helps to remember that the first dimension represents the respondent's propensity to be impacted by the features. As a result, we see the same rank ordering of the features repeated for each category level. For example, you can see the larger and darker red numbers decrease from 9 to 1 as you move down from the upper left side. Then, that pattern is repeated for the lighter and smaller red numbers, although Features 6 and 1 seem to be a little out of place. Feature 2 is hidden, but the light blue features are ordered as expected. Finally, the last repetition is for the larger dark blue numbers, even if Feature 4 moved out of line. In this example with simulated data, the features and the categories are well-separated. While we will always expect to see the categories for a single feature to be ordered, it is common to see overlap between different features and their respective categories.
It is worth our time to understand the arrangement of feature levels by examining the coordinates for the 36 columns that are plotted in the above graph (shown below). First, the four levels for each feature follow the same pattern with 1 < 2 < 3 < 4. Of course, this is what one would expect given that the scale was constructed to be ordinal. Still, we have passed a test since MCA does not force this ordering. The data entering a MCA are factors or nominal variables that are not ordered. Second, the fact that Feature 9 is preferred over Feature 1 can be seen in the placement of the four levels for each feature. Feature 1 starts at -0.39 (V1_1) and ends at 2.02 (V1_4). Feature 9, on the other hand, begins at -1.69 (V9_1) and finishes at 0.44 (V9_4). V9_1 has the lowest value on the first dimension because only a respondent with no latent interest in any feature would give the lowest score to the best feature. Similarly, the highest value on the first dimension belongs to V1_4 since only a zealot would pay more for the worst feature.
| Dim 1 | Dim 2 |
V1_1 | -0.39 | -0.06 |
V1_2 | 0.17 | -0.36 |
V1_3 | 1.06 | 0.91 |
V1_4 | 2.02 | 1.97 |
V2_1 | -0.58 | 0.13 |
V2_2 | 0.27 | -0.64 |
V2_3 | 0.96 | 0.55 |
V2_4 | 1.46 | 1.42 |
V3_1 | -0.80 | 0.13 |
V3_2 | 0.16 | -0.33 |
V3_3 | 0.86 | 0.03 |
V3_4 | 1.14 | 1.16 |
V4_1 | -0.97 | 0.18 |
V4_2 | -0.18 | 0.07 |
V4_3 | 0.41 | -0.39 |
V4_4 | 0.92 | 0.42 |
V5_1 | -0.88 | 0.42 |
V5_2 | -0.55 | -0.25 |
V5_3 | 0.24 | -0.54 |
V5_4 | 1.03 | 0.68 |
V6_1 | -1.26 | 0.68 |
V6_2 | -0.22 | -0.36 |
V6_3 | 0.08 | -0.41 |
V6_4 | 0.96 | 0.54 |
V7_1 | -1.54 | 1.28 |
V7_2 | -0.72 | -0.32 |
V7_3 | 0.04 | -0.36 |
V7_4 | 0.60 | 0.19 |
V8_1 | -1.52 | 1.59 |
V8_2 | -0.93 | -0.25 |
V8_3 | -0.20 | -0.25 |
V8_4 | 0.58 | 0.10 |
V9_1 | -1.69 | 2.24 |
V9_2 | -1.33 | 0.81 |
V9_3 | -0.30 | -0.34 |
V9_4 | 0.44 | -0.07 |
Finally, let us see how the respondents are positioned on the same MCA map. The red triangles are the 36 columns of the indicator matrix whose coordinates we have already seen in the above table. The blue dots are respondents. Respondents with similar response profiles are placed near each other. Given the feature structure shown in the heatmap, total score becomes a surrogate for respondent similarity. Defining a respondent's total score as the sum of the nine feature scores means that the total scores can range from 9 to 36. The 9'ers can be found at the top of the heatmap, and the 36'ers are at the bottom. It should be obvious that the closer the rows in the heatmap, the more similar the respondents.
Again, we see an arc that can be interpreted as the
manifold or
principal curve showing the trajectory of the underlying latent trait. The second dimension is a quadratic function of the first dimension (dim 2 = f(dim 1^2) with R-square = 0.86). This effect has been named the "horseshoe" effect. Although that name is descriptive, it encourages us to think of the arc as an artifact rather than a quadratic curve representing a scaling of the latent trait.
Finally, respondents fall along this arc in same order as their total scores. Respondent at the low end of the arc in the upper left are those giving the lowest scores to all the items. At the end of the arc in the upper right is where we find those respondents giving the highest feature scores.
Caveats and Other Conditions, Warnings and StipulationsEverything we have done depends on the respondent's ability to know and report accurately on how the additional feature will impact them in the marketplace. Self-report, however, does not have a good track record, though sometimes it is the best we can do when there are many features to be screened. Besides social desirability, the most serious limitation is that respondents tend to be too optimistic because their mental simulations do not anticipate the impediments that will occur when the product with the added feature is actually offered in the marketplace.
Prospection is no more accurate than retrospection.
Finally, it is an empirical question whether the graded response model captures the underlying feature prioritization process. To be clear, we are assuming that our behaviorally-anchored ratings are generated on the basis of a single continuous latent variable along which both the features and the respondents can be located. This may not be the case. Instead, we may have a multidimensional feature space, or our respondents may be a mixture of customer segments with different feature prioritization.
If customer heterogeneity is substantial and the product supports varying product configurations, we might find different segments wanting different feature sets. For example, feature bundles can be created to appeal to diverse customer segments as when a cable or direct TV provider offers sports programming or movie channel packages at a discounted price. However, this is not a simple finite mixture or latent class model but a hybrid mixture of customer types and intensities for some buyers of sports programming are sports fanatics who cannot live without it and other buyers are far less committed. You can read more about hybrid mixtures of categorical and continuous latent variables in a
previous post.
In practice, one must always be open to the possibility that your data set is not homogeneous but contains one or more segments seeking different features. Wants and needs are not like perceptions, which seem to be shared even by individuals with very different needs (e.g., you may not have children and never go to McDonald's but you know that it offers fast food for kids). Nonetheless, when the feature set does not contain bundles deliberately chosen to appeal to different segments, the graded response model seems to perform well.
I am aware that our model imposes a good deal of structure on the response generation process. Yet in the end, the graded response model reflects the feature prioritization process in the same way that a conjoint model reflects the choice process. Conjoint models assume that the product is a bundle of attributes and that product choice can be modeled as an additive combination of the value of attribute levels. If the conjoint model can predict choice, it is accepted as an "as if" model even when we do not believe that consumers stored large volumes of attribute values that they retrieve from memory and add together. They just behave as if they did.
Appendix: All the R code needed to create the data and run the analyses
I used a function from the psych library to generate the 4-point rating scale for the graded response model. One needs to set the difficulty values using d and realize that the more difficult items are the less popular ones (i.e., a "hard" feature finds it "hard" to have an impact, while an "easy" feature finds it "easy"). The function sim.poly.npl() need to know the number of variables(nvar), the number of respondents(n), the number of categories (cat), and the mean(mu) plus standard deviation (sd) for the normal distribution describing the latent trait differentiating among our respondents. The other parameters can be ignored for this example. The function returns a list with scale scores in bar$items from 0 to 3 (thus the +1 to get a 4-point scale from1 to 4).
The function heatmap.2() comes from the gplots package. Since I have sorted the data matrix by row and column marginals, I have suppressed the clustering of row and columns.
The MCA() function from FactoMineR needs factors, so there are three line showing how to make ratings a data frame and then use lapply to convert the numeric to factors. You will notice that I needed to flip the first dimension to run from low to high, so there are a number of lines that reverse the sign of the first dimension.
library(psych)
d<-c(1.50,1.25,1.00,.25,0,
-.25,-1.00,-1.25,-1.50)
set.seed(12413)
bar<-sim.poly.npl(nvar = 9, n = 200,
low=-1, high=1, a=NULL,
c=0, z=1, d=d, mu=0,
sd=1, cat=4)
ratings<-bar$items+1
library(gplots)
feature<-apply(ratings,2,mean)
person<-apply(ratings,1,sum)
ratingsOrd<-ratings[order(person),
order(feature)]
heatmap.2(as.matrix(ratingsOrd), Rowv=FALSE,
Colv=FALSE, dendrogram="none",
col=redblue(16), key=FALSE,
keysize=1.5, density.info="none",
trace="none", labRow=NA)
F.ratings<-data.frame(ratings)
F.ratings[]<-lapply(F.ratings, factor)
str(F.ratings)
library(FactoMineR)
mca<-MCA(F.ratings)
summary(mca)
categories<-mca$var$coord[,1:2]
categories[,1]<--categories[,1]
categories
feature_label<-c(rep(1,4),rep(2,4),rep(3,4),
rep(4,4),rep(5,4),rep(6,4),
rep(7,4),rep(8,4),rep(9,4))
category_color<-rep(c("darkred","red",
"blue","darkblue"),9)
category_size<-rep(c(1.1,1,1,1.1),9)
plot(categories, type="n")
text(categories, labels=feature_label,
col=category_color, cex=category_size)
mca2<-mca
mca2$var$coord[,1]<--mca$var$coord[,1]
mca2$ind$coord[,1]<--mca$ind$coord[,1]
plot(mca2, choix="ind", label="none")
R-bloggers.com offers
daily e-mail updates about
R news and
tutorials on topics such as: visualization (
ggplot2,
Boxplots,
maps,
animation), programming (
RStudio,
Sweave,
LaTeX,
SQL,
Eclipse,
git,
hadoop,
Web Scraping) statistics (
regression,
PCA,
time series,
trading) and more...